Make a precision-like matrix block-banded according to group membership, keeping only entries within specified group neighborhoods.
Arguments
- mat
A \(d \times d\) precision-like matrix specifying the base matrix to be masked.
- membership
An integer vector specifying the group membership. The length of
membershipmust be consistent with the dimension \(d\).- neighbor.range
An integer (default = 1) specifying the neighbor range, where groups whose labels differ by at most
neighbor.rangeare considered neighbors and kept in the mask.
Value
An object with S3 class "sparsify_block_banded" containing
the following components:
- Omega
The masked precision matrix.
- Sigma
The covariance matrix, i.e., the inverse of
Omega.- sparsity
Proportion of zero entries in
Omega.- membership
An integer vector specifying the group membership.
Examples
library(grasps)
## reproducibility for everything
set.seed(1234)
## precision matrix estimation
X <- matrix(rnorm(200), 10, 20)
membership <- c(rep(1,5), rep(2,5), rep(3,4), rep(4,6))
est <- grasps(X, membership = membership, penalty = "lasso", crit = "BIC")
## default: keep blocks within ±1 of each group
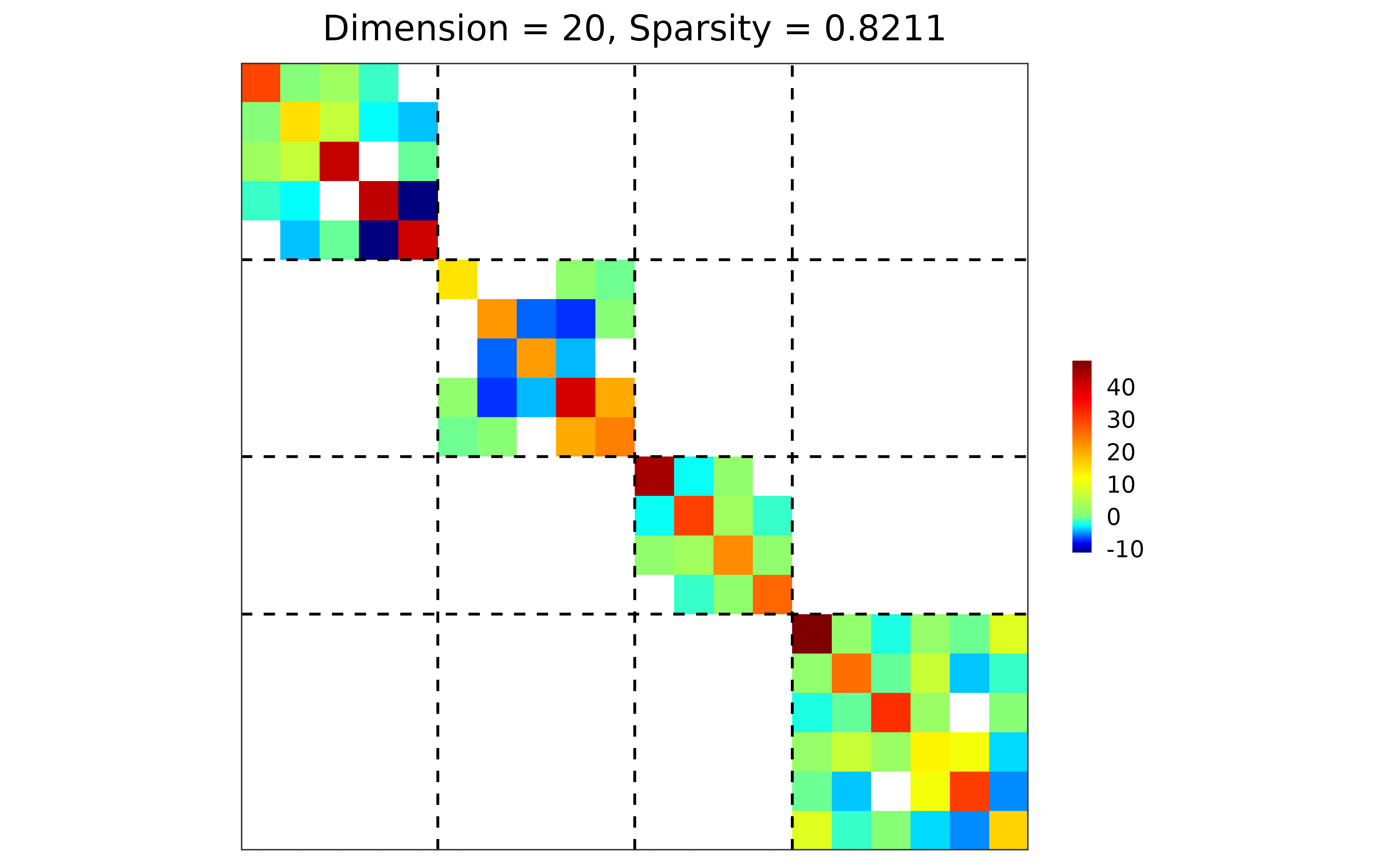
res1 <- sparsify_block_banded(est$hatOmega, membership, neighbor.range = 1)
plot(res1)
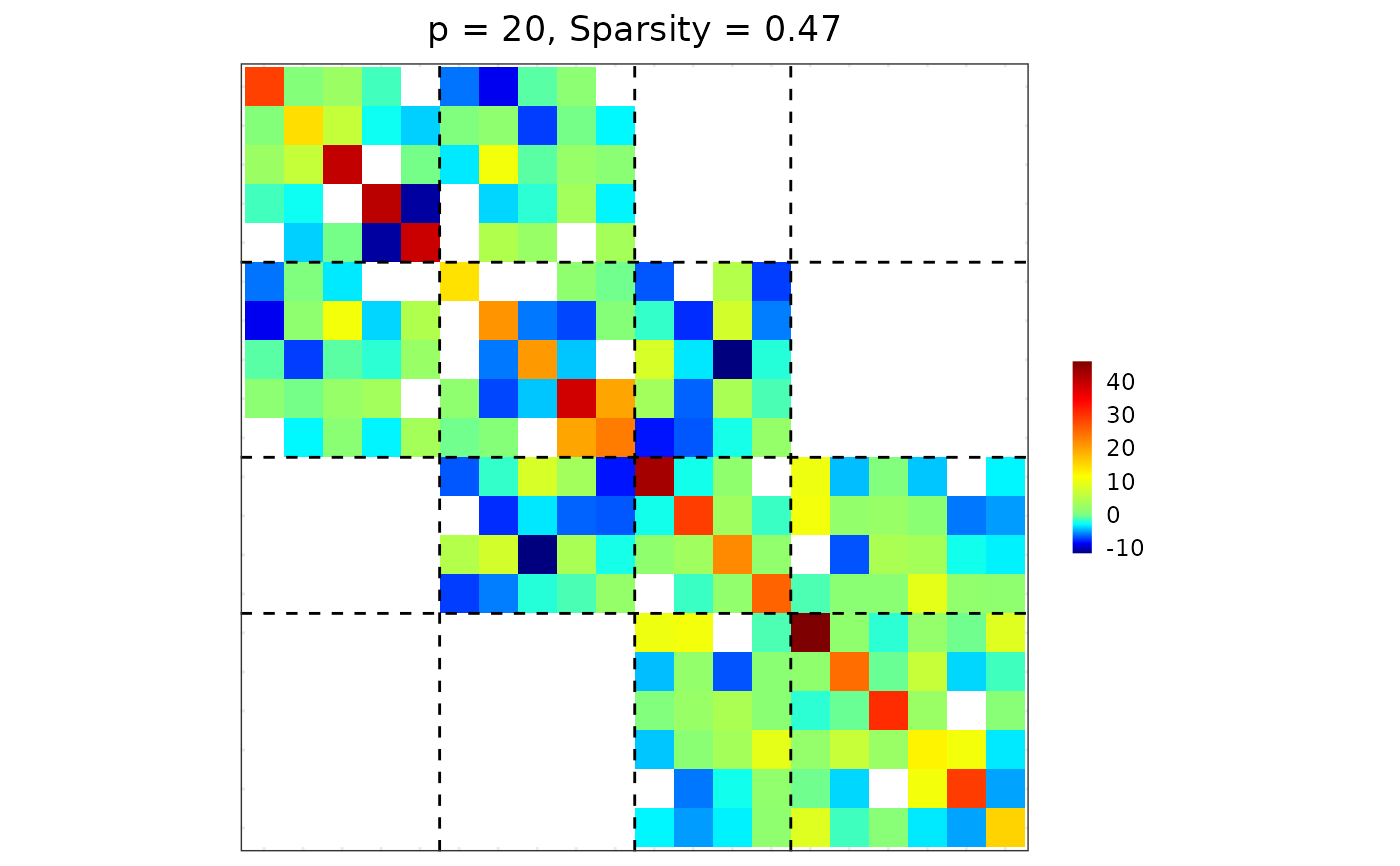 ## wider band: keep blocks within ±2 of each group
res2 <- sparsify_block_banded(est$hatOmega, membership, neighbor.range = 2)
plot(res2)
## wider band: keep blocks within ±2 of each group
res2 <- sparsify_block_banded(est$hatOmega, membership, neighbor.range = 2)
plot(res2)
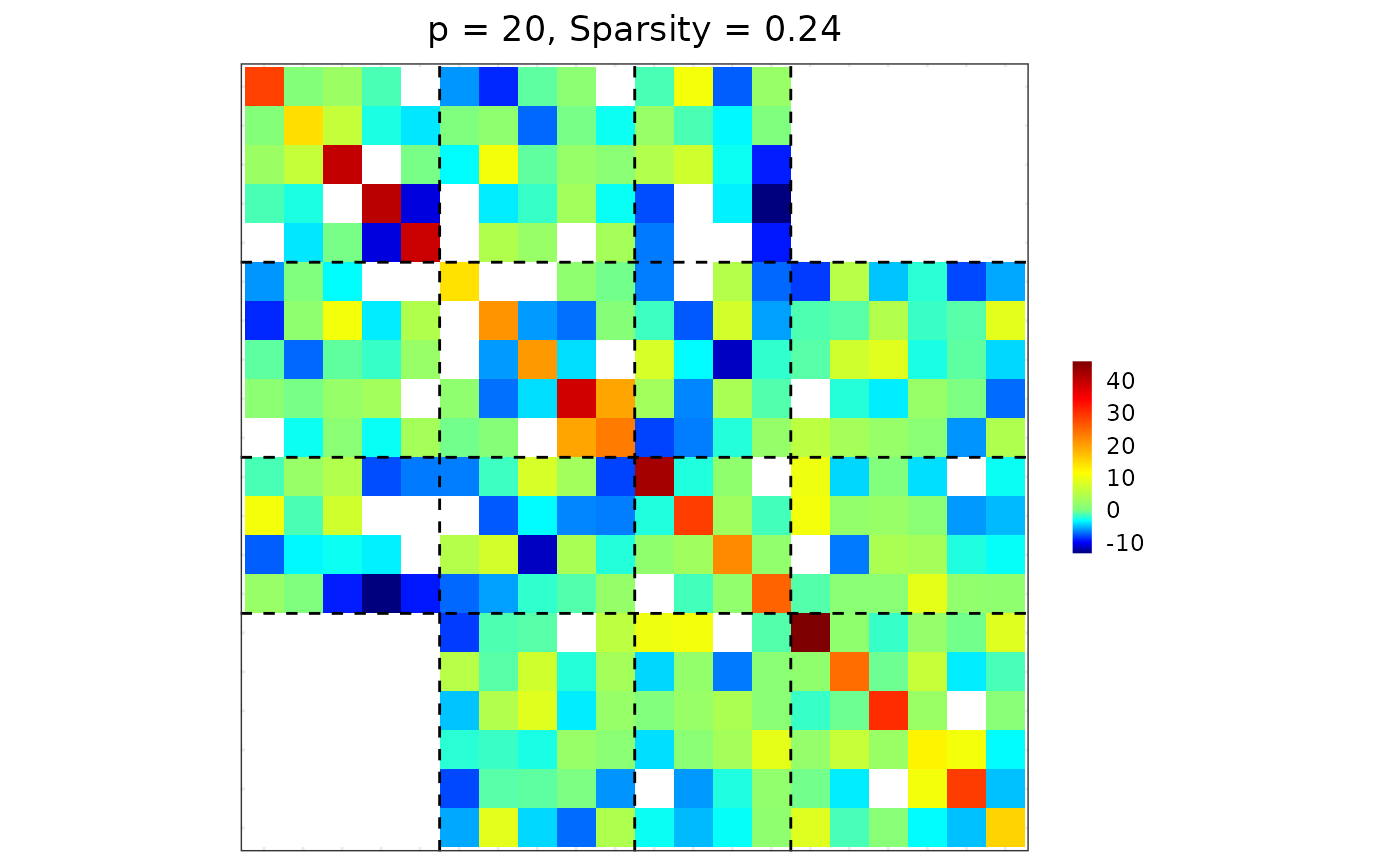 ## special case: block-diagonal matrix
res3 <- sparsify_block_banded(est$hatOmega, membership, neighbor.range = 0)
plot(res3)
## special case: block-diagonal matrix
res3 <- sparsify_block_banded(est$hatOmega, membership, neighbor.range = 0)
plot(res3)