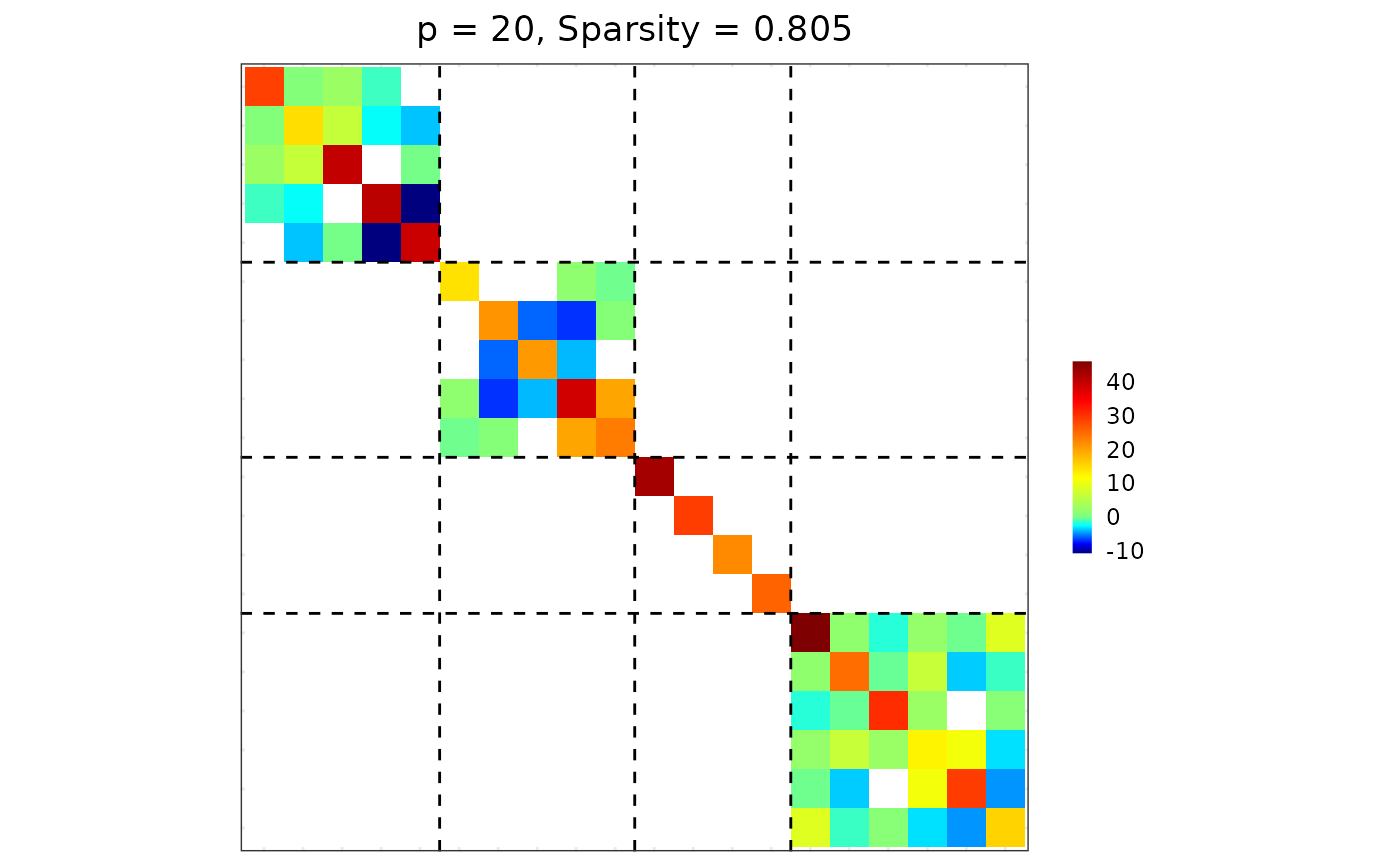
Make a precision-like matrix block-diagonal according to group membership, keeping only within-group blocks. Optionally, further reduce selected within-group blocks to diagonal-only (identity-like) structure.
Arguments
- mat
A p-by-p precision-like matrix specifying the base matrix to be masked.
- membership
An integer vector specifying the group membership. The length of
membershipmust be consistent with the dimension p.- group.diag
(default =
NULL)NULL: Makematblock-diagonal, i.e., only keep all within-group blocks.An integer vector specifying which within-group blocks are further reduced to diagonal-only structure (based on the block-diagonal form).
Value
A list containing:
- Omega
The masked precision matrix.
- Sigma
The covariance matrix, i.e., the inverse of
Omega.- sparsity
Proportion of zero entries in
Omega.- membership
An integer vector specifying the group membership.
Examples
## reproducibility for everything
set.seed(1234)
## precision matrix estimation
X <- matrix(rnorm(200), 10, 20)
membership <- c(rep(1,5), rep(2,5), rep(3,4), rep(4,6))
est <- grasps(X, membership = membership, penalty = "lasso", crit = "BIC")
## block-diagonalizel; within-group block for group 3 made diagonal-only.
res <- sparsify_block_diag(est$hatOmega, membership, group.diag = 3)
## visualization
visualize(res$Omega, res$membership)